Bruker Expands Proteomics Capabilities with New Software at US HUPO
At the 21st Annual US HUPO Conference, Bruker Corporation (Nasdaq: BRKR) announced significant advancements in 4D-Proteomics™ performance, software, and applications to enhance biological insights. These new developments aim to refine single-cell proteomics, immunopeptidomics, and glycoproteomics while integrating novel software tools for improved analysis and data interpretation.
A. AIP on timsTOF Ultra 2: Advancing Single-Cell Proteomics and Immunopeptidomics
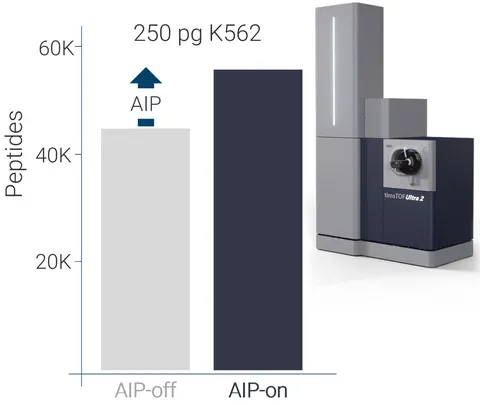
Bruker has introduced the new Advanced Ion Processing (AIP) technology on its timsTOF Ultra 2 mass spectrometer, delivering a substantial enhancement in peptide and protein identification. This work-in-progress innovation is designed to optimize ion transfer from the collision cell to the TOF analyzer, significantly improving identification capabilities in single-cell proteomics and immunopeptidomics.

For SCP-relevant samples containing 250 pg of material, AIP increases protein identifications by over 15% and peptide identifications by more than 20%, ensuring higher sequence coverage. Immunopeptidomics analysis sees similar improvements, enabling a more precise characterization of peptide fragments involved in immune responses.
Bruker plans to officially launch AIP on the timsTOF Ultra 2 system at ASMS 2025, with field upgradability for existing users.
B. DeutEx: Advancing Hydrogen-Deuterium Exchange (HDX)-MS Analysis
Bruker has also integrated DeutEx, a specialized software for analyzing hydrogen-deuterium exchange mass spectrometry (HDX-MS), into its proteomics ecosystem. Developed by Petr Novák from the Czech Academy of Sciences, DeutEx enhances the structural and dynamic study of proteins and complexes.
Dr. Petr Novák, Head of Structural Biology and Cell Signaling at the Institute of Microbiology of the Czech Academy of Sciences in Prague, emphasized the significance of this integration: “We are pleased that Bruker is integrating DeutEx for the analysis of HDX-MS. With support and further development by Bruker, DeutEx will become widely used to enhance the study of structural and dynamic properties of proteins and complexes, providing valuable insights for biopharma research and structural biology.”
By supporting timsTOF, MRMS, and MALDI-TOF systems, DeutEx offers researchers powerful tools to elucidate protein interactions, conformational changes, and the effects of ligand binding.
C. OmniScape 2025b: Next-Generation Integrative Top-Down Proteomics
Bruker has released OmniScape 2025b, an upgraded version of its integrative top-down proteomics software. This latest iteration delivers a tenfold increase in de novo sequencing performance, enhanced detection of truncated protein variants, and comprehensive sequence annotation.
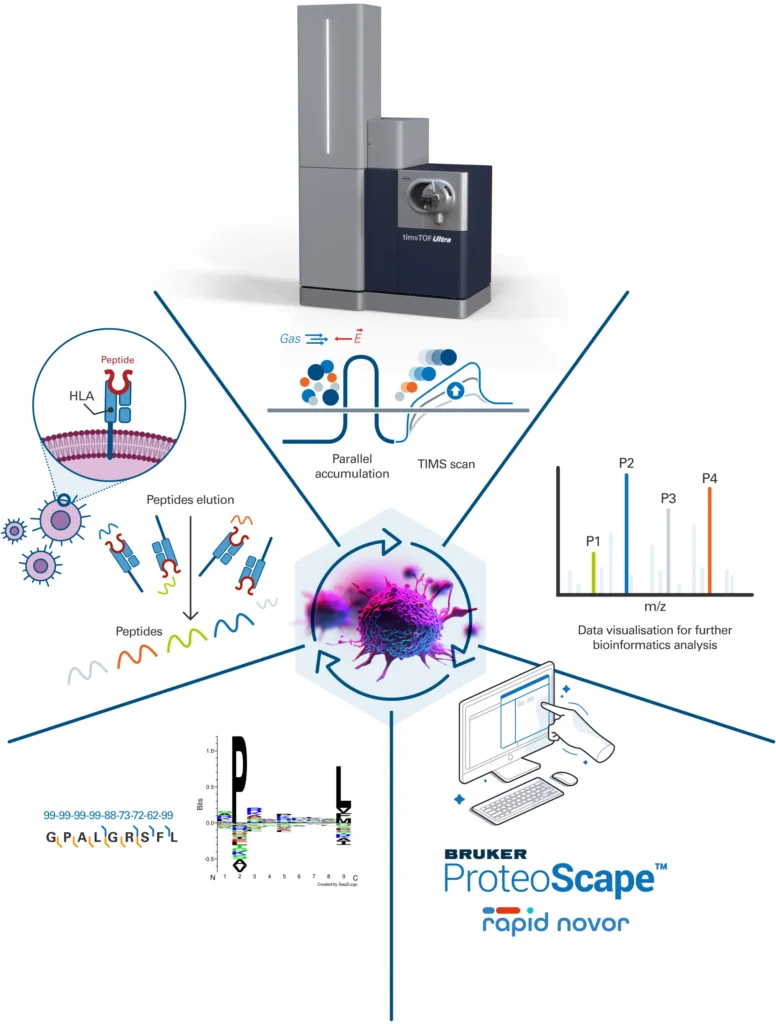
OmniScape 2025b supports the integration of sequence maps from various MSn levels (MS2 and MS3), including MALDI T³-sequencing, offering a multi-faceted approach to proteoform characterization.
Professor Julia Chamot-Rooke of the Institut Pasteur in Paris praised the software’s capabilities: “OmniScape is the most advanced software in my lab for analyzing top-down MS/MS protein spectra from any kind of fragmentation method.”
Professor Francisco Alberto Fernandez-Lima of Florida International University also highlighted its effectiveness: “Our lab has extensively utilized OmniScape for de novo and top-down protein-PTM sequence validation from ExD and UVPD datasets from timsTOF and FT-ICR MS instruments. Unlike alternative approaches, OmniScape is user-friendly and allows interactive fragment assignment/confirmation of proteoforms, including variable amino acid-level PTMs, m/z internal calibration, and MS-BLAST search for database inquiries.”
D. Enhanced ProteoScape Processing and Scalable Glycoproteomics Insights
Bruker’s ProteoScape™ V.2025c offers notable updates, including Spectronaut® v.19.6 and GlycoScape™ v.2025c, improving proteomics and glycoproteomics analysis.
Spectronaut v.19.6 introduces performance enhancements such as improved quantity imputation and PTM stoichiometry calculations. Meanwhile, GlycoScape v.2025c now allows users to customize modifications, including glycan building block selection and labeled glycans.
When used alongside Agilent AssayMAP Bravo®, GlycoScape v.2025c facilitates automated, high-throughput glycan profiling. This advancement results in a 6x-10x increase in glycopeptide identifications, enabling large-scale glycoform characterization on timsTOF systems with real-time GlycoScape processing.
Dr. Christopher Adams, Director of Proteomics at Rezo Therapeutics, expressed his satisfaction with the ProteoScape platform: “Our timsTOF HT has been synchronized with the ProteoScape platform, and I am impressed by the speed in providing new versions and capabilities.”
E. TwinScape: AI-Enabled Cloud-Based Proteomics Quality Monitoring
Bruker has introduced TwinScape, an AI-enabled, cloud-based quality monitoring software that provides real-time system diagnostics for proteomics researchers. TwinScape is currently being used to monitor hundreds of customer systems, including nanoElute® LCs and the entire timsTOF series.
Dr. Julian Langer, Head of the Mass Spectrometry Lab at the Max Planck Institute of Biophysics and Brain Research in Frankfurt, Germany, shared his experience: “TwinScape has been a great addition to our HeLa/iRT-based quality control pipeline and is part of our routine QC checks. The level of insight and constant monitoring that TwinScape provides has dramatically improved our workflow efficiency. Not only has it allowed us to address potential problems before they escalate, but it has also enabled proactive technical customer support.